Help for M-DeepAssembly2 server
Input of M-DeepAssembly2 server
Static and multiple conformations prediction:
-
(Mandatory) At least 1 full-chain protein sequence in Fasta format in PDB format
, or at least 2 structure models in PDB format for multiple domain proteins.
Users can click the button "Add model" to add text boxes for input more domain structure models of multiple domain proteins. The server page is set to receive up to 10 domain structure models. - If you choose to upload a file containing a multi-domain protein structure, please ensure that each model file ends in ".pdb".
- (Mandatory) Provide your email address
- (Optional) Assign your job name
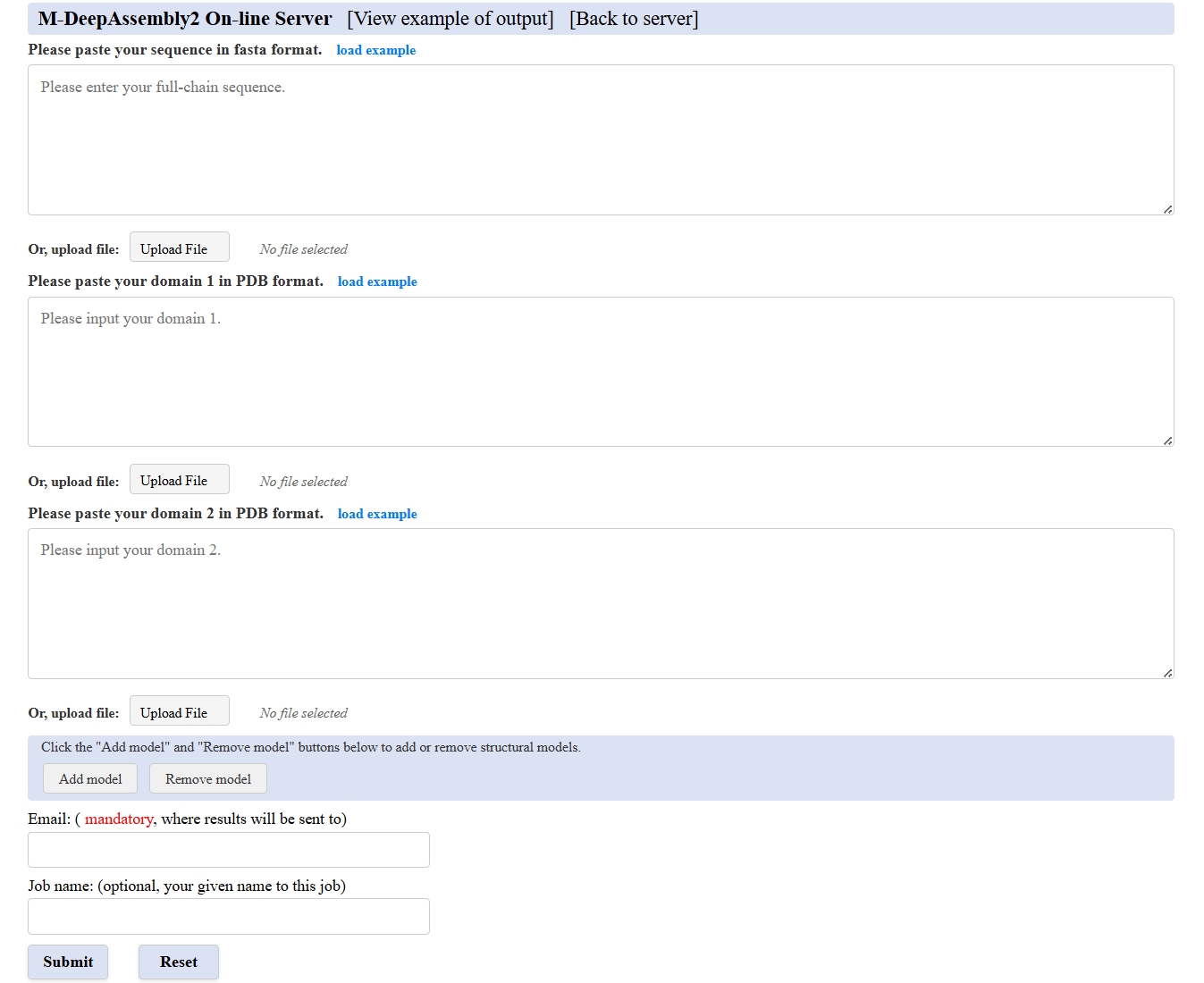 Figure 1. The "Submit" section of multiple conformations prediction in M-DeepAssembly2 home page.
Figure 1. The "Submit" section of multiple conformations prediction in M-DeepAssembly2 home page.
Output of M-DeepAssembly2 server
Protein multiple conformations prediction:
- The multiple conformations of input protein.
- The results page shows the 12 models randomly selected from all predicted results, The complete set of predictions can be downloaded by clicking on the pdb.pdb above.
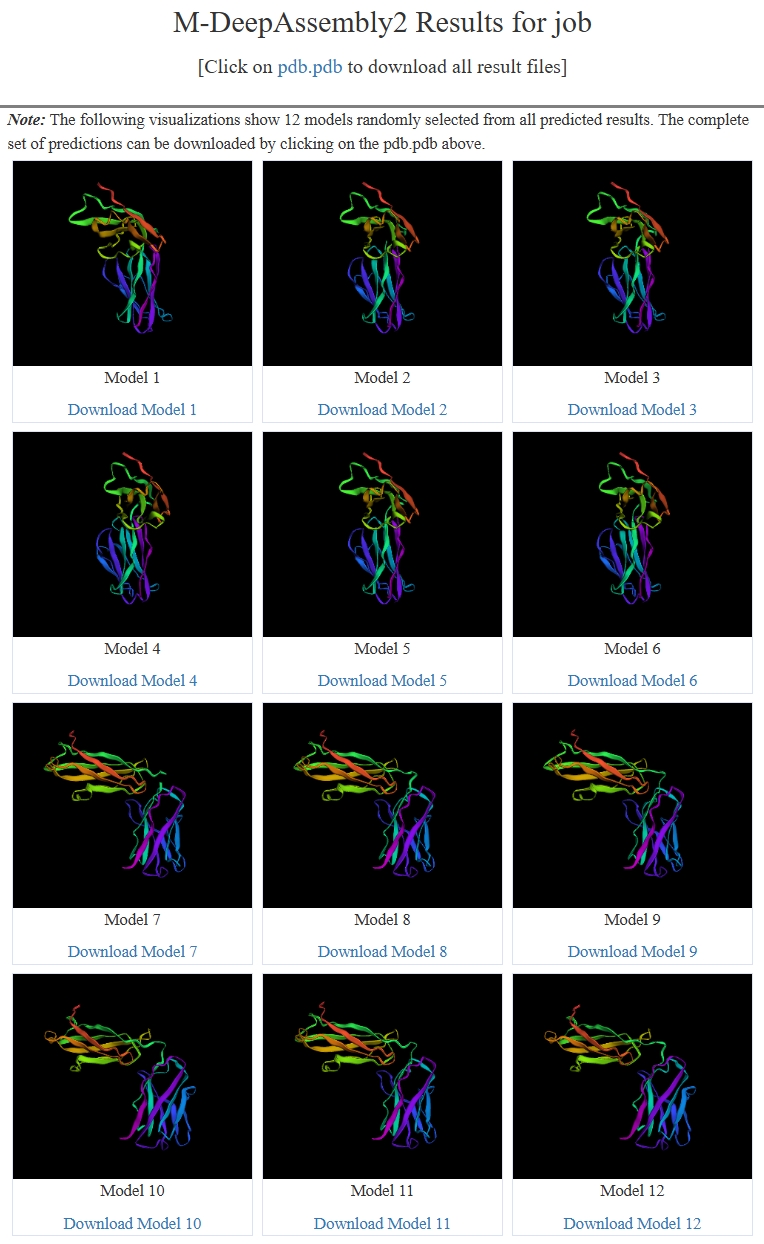 Figure 2. The "Submit" section of multiple conformations prediction in M-DeepAssembly2 home page.
Figure 2. The "Submit" section of multiple conformations prediction in M-DeepAssembly2 home page.
How to cite M-DeepAssembly2?
Please cite the following articles when you use the M-DeepAssembly2 server:
- Xinyue Cui✝, Yuhao Xia✝, Minghua Hou, Xuanfeng Zhao, Suhui Wang and Guijun Zhang*. M-DeepAssembly:Enhanced DeepAssembly based on multi-objective protein conformation sampling. BMC bioinformatics, 26, 120 (2025). DOI: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-025-06131-2.
- Yuhao Xia✝, Kailong Zhao✝, Dong Liu, Xiaogen Zhou, Guijun Zhang*. AlphaFlex: Accuracy modeling of protein multiple conformations via predicted flexible residues. Communications biology, 6, 1221 (2023). DOI: https://www.nature.com/articles/s42003-023-05610-7.
- Lingyu Ge✝, Xinyue Cui✝, Kailong Zhao, Xiaogen Zhou, Yang Zhang, Guijun Zhang*. Multi-domain and complex protein structure prediction using inter-domain interactions from deep learning. bioRxiv, 2025-07 (2023). DOI: https://www.biorxiv.org/content/10.1101/2025.07.11.664327v1.
- Xinyue Cui✝, Lingyu Ge✝, Xia Chen, Zexin Lv, Suhui Wang, Xiaogen Zhou, Guijun Zhang*. Beyond static structures: protein dynamic conformations modeling in the post-AlphaFold era. Briefings in bioinformatics, 26, 4 (2025). DOI: https://academic.oup.com/bib/article/26/4/bbaf340/8202937.
Need more help?
If you have more questions or comments about the server, please email Cuixinyue@zjut.edu.cn.