Help for DeepUMQA-X server
Input of DeepUMQA-X server
Protein single-chain structure assessment:
-
(Mandatory) At least 1 protein single-chain structure model in PDB format
Users can click the button "Add model" to add text boxes for input more structure models of same protein single-chain. The server page is set to receive up to 10 structure models. - If you choose to upload a zip file containing all models of the same protein (each model file ends with ".pdb"), please compress the folder to ./files or ./dir/files.
- Specifies the evaluation method type. If you specify Single-model method, the input model will be evaluated using GraphCPLMQA2. If you specify Consensus method, the input model will be evaluated using GraphCPLMQA2 with lightweight structural alignment. This result contains relative information between different structural models. Consensus method is not recommended when the number of models is less than 50.
- (Optional) Provide your email address
- (Optional) Assign your job name
 Figure 1. The "Submit" section of single-chain structure assessment in DeepUMQA-X home page.
Figure 1. The "Submit" section of single-chain structure assessment in DeepUMQA-X home page.
-
(Mandatory) At least 1 protein complex structure model in PDB format
Users can click the button "Add model" to add text boxes for input more structure models of same protein complex The server page is set to receive up to 10 structure models. - If you choose to upload a zip file containing all models of the same protein (each model file ends with ".pdb"), please compress the folder to ./files or ./dir/files.
- Specifies the evaluation method type. If you specify Single-model method, the input model will be evaluated using GraphCPLMQA2. If you specify Consensus method, the input model will be evaluated using GraphCPLMQA2 with lightweight structural alignment. This result contains relative information between different structural models. Consensus method is not recommended when the number of models is less than .
- (Optional) Provide your email address
- (Optional) Assign your job name
 Figure 2. The "Submit" section of complex structure assessment in DeepUMQA-X home page.
Figure 2. The "Submit" section of complex structure assessment in DeepUMQA-X home page.
Output of DeepUMQA-X server
Protein single-chain structure assessment:
- The evaluated single-chain structure model
- Inter-residue distance deviation of the single-chain structure model
- TM-score and Global-lDDT of the single-chain structure model
- Per-residue lDDT of the single-chain structure model
- Ranking of all input single-chain structure model (According to the lDDT)
- A compressed package (.tar.gz) containing the above information.
- The results page shows the top 10 results, where the ranking is based on the Global-lDDT.Please note that if there are multiple large proteins, Jmol may not be able to display them due to memory limitations.
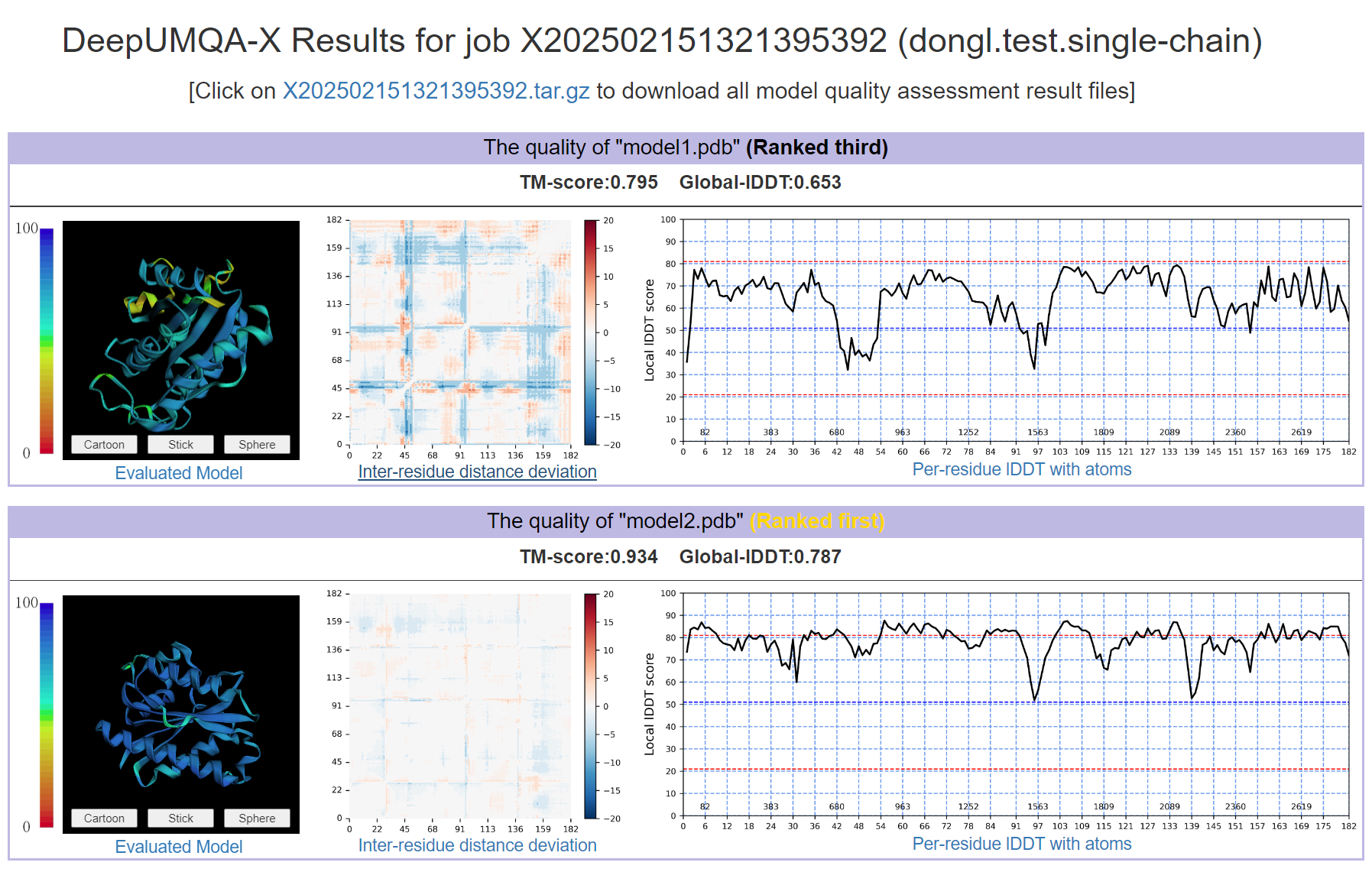 Figure 3. The output of single-chain structure assessment in DeepUMQA-X result page.
Figure 3. The output of single-chain structure assessment in DeepUMQA-X result page.
- The evaluated complex structure model
- TM-score, QS-score, Global-lDDT and Interface-lDDT of the complex structure model
- Per-residue lDDT of the complex structure model
-
Interface residues lDDT of the complex structure model
Interface residues are defined as having a contact with at least one residue from another chain (CB-CB distance ≤ 8Å, CA in case of Glycine) - A compressed package (.tar.gz) containing the above information.
- The results page shows the top 5 results, where the ranking is based on the Interface-lDDT.Please note that if there are multiple large proteins, Jmol may not be able to display them due to memory limitations.
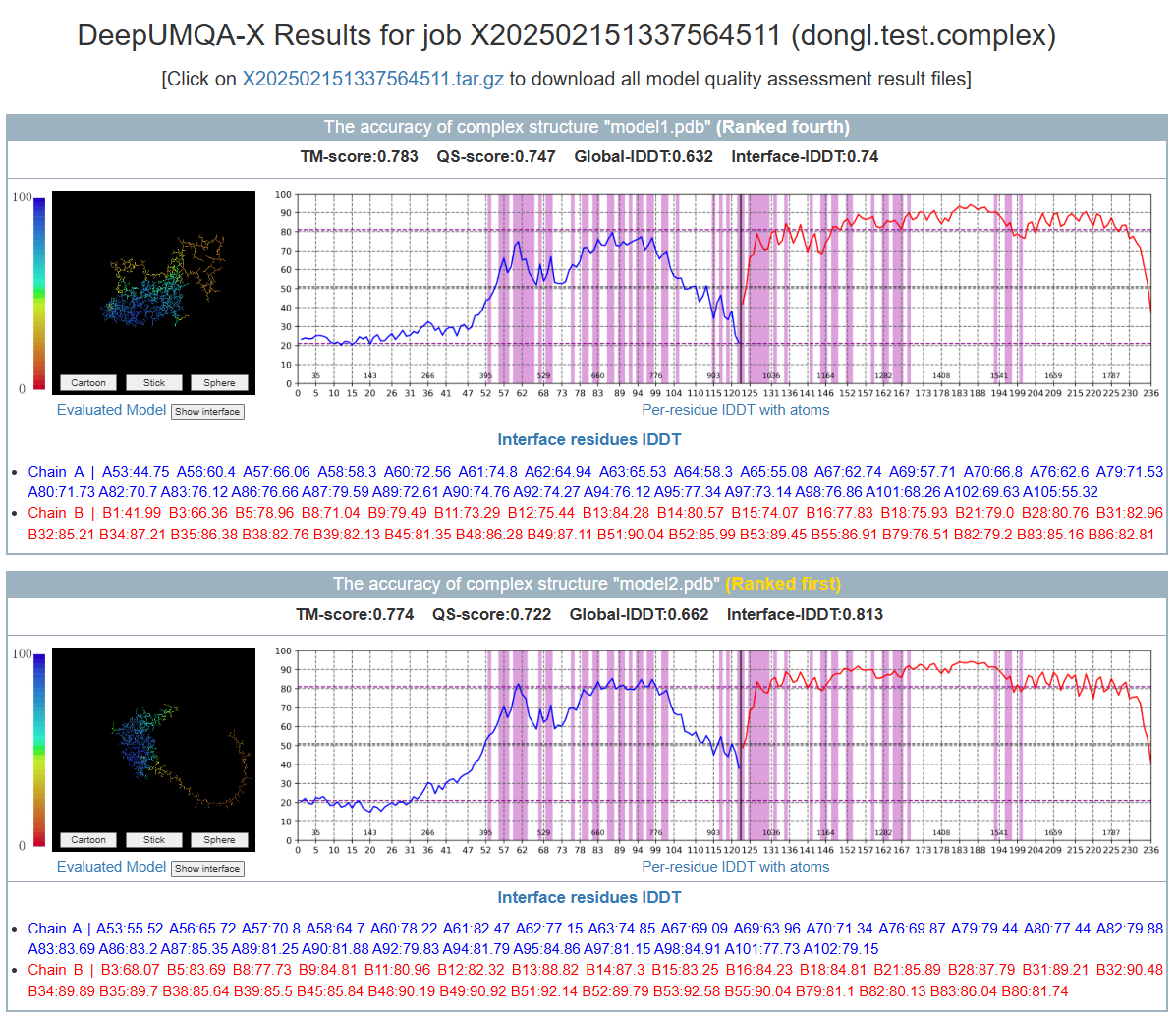 Figure 4. The output of complex structure assessment in DeepUMQA-X result page.
Figure 4. The output of complex structure assessment in DeepUMQA-X result page.
Evaluation Metrics
DeepUMQA-X uses three protein model evaluation metrics to describe its structural quality,
including TM-score (overall fold accuracy), QS-score (overall interface accuracy), and lDDT (local residues accuracy).
- TM-score (Zhang and Skolnick 2004) were calculated using US-align (Zhang et al. 2022) to assess protein single-chain and complex topological similarity.
- QS-score (Bertoni et al.) quantifies the similarity between interfaces based on shared interface contacts and is a global comparison of the entire complex.
- lDDT (Mariani et al.) assesses the differences in interatomic distances between model and reference structure where each interatomic distance ≤15Å in the reference structure is compared with its model structure. lDDT calculates the average of the distance difference scores below thresholds [0.5, 1.0, 2.0, 4.0].
How to cite DeepUMQA-X?
Please cite the following articles when you use the DeepUMQA-X server:
- Dong Liu✝, Biao Zhang✝, Jun Liu, Hui Li*, Le Song* and Guijun Zhang*. GraphCPLMQA: Assessing protein model quality based on deep graph coupled networks using protein language model. Briefings in Bioinformatics, 2023, 25(1):bbad420.
- Jun Liu, Kailong Zhao and Guijun Zhang*. Improved model quality assessment using sequence and structural information by enhanced deep neural networks. Briefings in Bioinformatics, 2023, 24(1): bbac507.
- Saisai Guo✝, Jun Liu✝, Xiaogen Zhou, Guijun Zhang*. DeepUMQA: Ultrafast Shape Recognition-based Protein Model Quality Assessment using Deep Learning. Bioinformatics, 2022, 38(7): 1895-1903.
Need more help?
If you have more questions or comments about the server, please email guijunlab06@163.com.